The TCF/LEF family (T cell factor/lymphoid enhancer factor family) is a group of genes that encode transcription factors which bind to DNA through a SOX-like high mobility group domain. They are involved in the Wnt signaling pathway, particularly during embryonic [2] and stem-cell development, [3] but also had been found to play a role in cancer [4] and diabetes. [5] TCF/LEF factors recruit the coactivator beta-catenin to enhancer elements of genes they target. They can also recruit members of the Groucho family of corepressors. [6]
History
The discovery of the TCF/LEF genes as nuclear Wnt pathway components in the 90s [7] [8] was a pivotal breakthrough for the Wnt signalling research field, plugging an important knowledge gap and enabling subsequent understanding of transcriptional regulation of Wnt target genes, particularly in embryonic development and cancer.
Before this discovery it was only known that upstream Wnt signalling mechanisms regulated the cytoplasmic abundance of the beta-catenin protein, which as a consequence translocated into the cell nucleus. However, since the protein structure of beta-catenin did not reveal any DNA-binding domain, it was still unclear how nuclear beta-catenin could regulate Wnt target genes. Following the discovery, a model was established whereby Wnt signalling-regulated beta-catenin in the nucleus attaches to TCF/LEF DNA binding proteins, which recognise the DNA consensus sequence around the core 'CTTTG', called Wnt Response Element (WRE). [9]
This rule that beta-catenin-TCF interaction on DNA regulates Wnt target gene expression, has nonetheless been broken by examples of Wnt- and beta-catenin-independent functions for TCF/LEF proteins (for instance in zebrafish CNS development [10]) and functional association of Wnt-regulated beta-catenin with other DNA-binding transcription factors such as SOX, [11] FOXO, [12] TBX. [13] Then again, this beta-catenin-TCF interaction on DNA is now revealed as but the core of much larger protein complexes regulating transcription, called the Wnt enhanceosomes. [14] Conversely, additional mechanisms regulating TCF/LEF protein function have been discovered, such as phosphorylation [15] and sumoylation. [16]
Structure
The structure and function of TCF/LEF proteins explains this bimodal function. TCF/LEF genes encode proteins with an elaborate structure that can however be summarised by considering four main domains:
- N-terminal domain: mediating interaction with beta-catenin, which is highly conserved and mediates the transcriptional activator function.
- Control region: includes sequences regulating and mediating the transcriptional repressor function and encoding a transcriptional repressor binding domain for the Groucho family.
- DNA-binding domain: includes a very highly conserved HMG ( High Mobility Group) DNA-binding domain and the NLS ( nuclear localisation sequence).
- C-terminal tail: may contain an additional DNA-binding domain and an additional transcriptional repressor binding domain. [17]
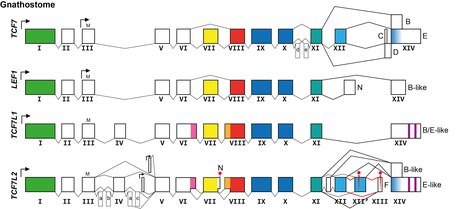
Diversity in TCF/LEF protein structure and function comes from having different genes. Humans and jawed vertebrates generally have four genes encoding TCF/LEF proteins:
- TCF7 (also called TCF1)
- LEF1 (also called TCF1α)
- TCF7L1 (also called TCF3)
- TCF7L2 (also called TCF4)
Further diversity comes from expression from the same gene of alternative transcripts encoding different protein isoforms, particularly from the TCF7 and TCF7L2 genes:
- There are isoforms expressed from secondary promoters that encode proteins that lack the usual N-terminus and therefore specifically the beta-catenin binding domain (see above). These protein isoforms function not as bimodal transcription factors but as constitutive repressors, and they are refractory to upstream Wnt signalling regulation. [18] [19]
- There are isoforms from alternative splicing in the part of the transcript encoding the control region, which influence the propensity of the encoded protein isoforms to act as transcriptional repressors (without beta-catenin) or transcriptional activators (with beta-catenin). [20]
- There are isoforms from alternative splicing in the part of the transcript encoding the C-terminal tail resulting in protein isoforms with and without the additional DNA-binding domain and resulting in changing the reading frame in which the last exon is translated with or without an additional transcriptional co-repressor binding domain. [21] [22]
Function
TCF/LEF proteins function as bimodal transcription factors:
- As described, TCF/LEF proteins act as transcriptional activators in association with nuclear beta-catenin (and transcriptional co-activators attached to beta-catenin);
- but without beta-catenin, TCF/LEF proteins function as transcriptional repressors (attached to transcriptional co-repressors members of the Groucho family).
Thus, as a consequence, Wnt target genes are actively repressed in the absence of Wnt signalling activity, then activated when Wnt signalling actively drives beta-catenin into the nucleus. [23]
TCF/LEF genes support diverse functions in embryonic development, stem cell biology, and in disease. [24] [25] Given the conservation of structure, functions of different TCF/LEF genes and proteins are often redundant in many organs and tissues where Wnt signalling is important, yet genetic analysis suggested from the beginning that this redundancy is only partial, suggesting TCF/LEF gene- and TCF isoform-specific functions, many of which are only now beginning to be discovered.
Prominent functions of TCF/LEF genes in embryonic development include vertebrate dorsal axis induction, anterior-posterior patterning of the developing Central Nervous System, neural crest development and many functions in organ development. Prominent functions of TCF/LEF genes in stem cell development have been particularly well dissected during the hair follicle cycle. [26] [27] TCF/LEF genes have roles in many cancers, with their role in colorectal cancer possibly being the best understood. [28] However, other human diseases have also been linked to TCF/LEF genes, particularly type 2 diabetes. [29] [30]
References
- ^ Torres‐Aguila, Nuria P.; Salonna, Marika; Hoppler, Stefan; Ferrier, David E. K. (2022-02-03). "Evolutionary diversification of the canonical Wnt signaling effector TCF/LEF in chordates". Development, Growth & Differentiation. 64 (3): 120–137. doi: 10.1111/dgd.12771. hdl: 2164/18617. ISSN 0012-1592. PMC 9303524. PMID 35048372. S2CID 246161317.
- ^ Logan, Catriona Y.; Nusse, Roel (November 2004). "The WNT Signaling Pathway in Development and Disease". Annual Review of Cell and Developmental Biology. 20 (1): 781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. PMID 15473860.
- ^ Nusse, R (May 2008). "Wnt signaling and stem cell control". Cell Research. 18 (5): 523–7. doi: 10.1038/cr.2008.47. PMID 18392048. S2CID 2503910.
- ^ Zhan, T; Rindtorff, N; Boutros, M (March 2017). "Wnt signaling in cancer". Oncogene. 36 (11): 1461–1473. doi: 10.1038/onc.2016.304. PMC 5357762. PMID 27617575.
- ^ Laudes, M (April 2011). "Role of WNT signalling in the determination of human mesenchymal stem cells into preadipocytes". Journal of Molecular Endocrinology. 46 (2): R65-72. doi: 10.1530/JME-10-0169. PMID 21247979.
- ^ Brantjes H, Barker N, van Es J, Clevers H (February 2002). "TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling". Biol. Chem. 383 (2): 255–61. doi: 10.1515/BC.2002.027. PMID 11934263. S2CID 25665021.
- ^ Behrens, J; von Kries, JP; Kühl, M; Bruhn, L; Wedlich, D; Grosschedl, R; Birchmeier, W (15 August 1996). "Functional interaction of beta-catenin with the transcription factor LEF-1". Nature. 382 (6592): 638–42. Bibcode: 1996Natur.382..638B. doi: 10.1038/382638a0. PMID 8757136. S2CID 4369341.
- ^ Huber, O; Korn, R; McLaughlin, J; Ohsugi, M; Herrmann, BG; Kemler, R (September 1996). "Nuclear localization of beta-catenin by interaction with transcription factor LEF-1". Mechanisms of Development. 59 (1): 3–10. doi: 10.1016/0925-4773(96)00597-7. PMID 8892228. S2CID 14324471.
- ^ Cadigan, KM; Waterman, ML (1 November 2012). "TCF/LEFs and Wnt signaling in the nucleus". Cold Spring Harbor Perspectives in Biology. 4 (11): a007906. doi: 10.1101/cshperspect.a007906. PMC 3536346. PMID 23024173.
- ^ Duncan, RN; Panahi, S; Piotrowski, T; Dorsky, RI (2015). "Identification of Wnt Genes Expressed in Neural Progenitor Zones during Zebrafish Brain Development". PLOS ONE. 10 (12): e0145810. Bibcode: 2015PLoSO..1045810D. doi: 10.1371/journal.pone.0145810. PMC 4699909. PMID 26713625.
- ^ Kormish, JD; Sinner, D; Zorn, AM (January 2010). "Interactions between SOX factors and Wnt/beta-catenin signaling in development and disease". Developmental Dynamics. 239 (1): 56–68. doi: 10.1002/dvdy.22046. PMC 3269784. PMID 19655378.
- ^ Essers, MA; de Vries-Smits, LM; Barker, N; Polderman, PE; Burgering, BM; Korswagen, HC (20 May 2005). "Functional interaction between beta-catenin and FOXO in oxidative stress signaling". Science. 308 (5725): 1181–4. Bibcode: 2005Sci...308.1181E. doi: 10.1126/science.1109083. PMID 15905404. S2CID 24572861.
- ^ Zimmerli, Dario; Borrelli, Costanza; Jauregi-Miguel, Amaia; Söderholm, Simon; Brütsch, Salome; Doumpas, Nikolaos; Reichmuth, Jan; Murphy-Seiler, Fabienne; Aguet, MIchel; Basler, Konrad; Moor, Andreas E; Cantù, Claudio (18 August 2020). "TBX3 acts as tissue-specific component of the Wnt/β-catenin transcriptional complex". eLife. 9. doi: 10.7554/eLife.58123. PMC 7434441. PMID 32808927.
- ^ Gammons, M; Bienz, M (April 2018). "Multiprotein complexes governing Wnt signal transduction". Current Opinion in Cell Biology. 51: 42–49. doi: 10.1016/j.ceb.2017.10.008. PMID 29153704.
- ^ Sokol, SY (July 2011). "Wnt signaling through T-cell factor phosphorylation". Cell Research. 21 (7): 1002–12. doi: 10.1038/cr.2011.86. PMC 3193496. PMID 21606952.
- ^ Yamamoto, H; Ihara, M; Matsuura, Y; Kikuchi, A (1 May 2003). "Sumoylation is involved in beta-catenin-dependent activation of Tcf-4". The EMBO Journal. 22 (9): 2047–59. doi: 10.1093/emboj/cdg204. PMC 156076. PMID 12727872.
- ^ Hoppler, Stefan; Waterman, Marian L. (2014). "Evolutionary Diversification of Vertebrate TCF/LEF Structure, Function, and Regulation". WNT Signaling in Development and Disease: 225–237. doi: 10.1002/9781118444122.ch17. ISBN 9781118444122.
- ^ Van de Wetering, M; Castrop, J; Korinek, V; Clevers, H (March 1996). "Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties". Molecular and Cellular Biology. 16 (3): 745–52. doi: 10.1128/MCB.16.3.745. PMC 231054. PMID 8622675.
- ^ Hovanes, K; Li, TW; Munguia, JE; Truong, T; Milovanovic, T; Lawrence Marsh, J; Holcombe, RF; Waterman, ML (May 2001). "Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer". Nature Genetics. 28 (1): 53–7. doi: 10.1038/ng0501-53. PMID 11326276. S2CID 28974522.
- ^ Liu, F; van den Broek, O; Destrée, O; Hoppler, S (December 2005). "Distinct roles for Xenopus Tcf/Lef genes in mediating specific responses to Wnt/beta-catenin signalling in mesoderm development". Development. 132 (24): 5375–85. doi: 10.1242/dev.02152. PMID 16291789. S2CID 20515922.
- ^ Atcha, FA; Syed, A; Wu, B; Hoverter, NP; Yokoyama, NN; Ting, JH; Munguia, JE; Mangalam, HJ; Marsh, JL; Waterman, ML (December 2007). "A unique DNA binding domain converts T-cell factors into strong Wnt effectors". Molecular and Cellular Biology. 27 (23): 8352–63. doi: 10.1128/MCB.02132-06. PMC 2169181. PMID 17893322.
- ^ Ravindranath, AJ; Cadigan, KM (3 August 2016). "The Role of the C-Clamp in Wnt-Related Colorectal Cancers". Cancers. 8 (8): 74. doi: 10.3390/cancers8080074. PMC 4999783. PMID 27527215.
- ^ Ramakrishnan, AB; Cadigan, KM (2017). "Wnt target genes and where to find them". F1000Research. 6: 746. doi: 10.12688/f1000research.11034.1. PMC 5464219. PMID 28649368.
- ^ Hoppler, S; Kavanagh, CL (1 February 2007). "Wnt signalling: variety at the core". Journal of Cell Science. 120 (Pt 3): 385–93. doi: 10.1242/jcs.03363. PMID 17251379. S2CID 30976795.
-
^ Hoppler, Stefan; Moon, Randall T. (2014). Wnt signaling in development and disease : molecular mechanisms and biological functions. Hoboken, New Jersey.
ISBN
9781118444122.
{{ cite book}}: CS1 maint: location missing publisher ( link) - ^ DasGupta, R; Fuchs, E (October 1999). "Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation". Development. 126 (20): 4557–68. doi: 10.1242/dev.126.20.4557. PMID 10498690.
- ^ Merrill, BJ; Gat, U; DasGupta, R; Fuchs, E (1 July 2001). "Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin". Genes & Development. 15 (13): 1688–705. doi: 10.1101/gad.891401. PMC 312726. PMID 11445543.
- ^ Mayer, Claus-Dieter; Magon de La Giclais, Soizick; Alsehly, Fozan; Hoppler, Stefan (May 2020). "Diverse LEF/TCF Expression in Human Colorectal Cancer Correlates with Altered Wnt-Regulated Transcriptome in a Meta-Analysis of Patient Biopsies". Genes. 11 (5): 538. doi: 10.3390/genes11050538. PMC 7288467. PMID 32403323.
- ^ Jin, T; Liu, L (November 2008). "The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus". Molecular Endocrinology. 22 (11): 2383–92. doi: 10.1210/me.2008-0135. PMID 18599616.
- ^ Chen, X; Ayala, I; Shannon, C; Fourcaudot, M; Acharya, NK; Jenkinson, CP; Heikkinen, S; Norton, L (April 2018). "The Diabetes Gene and Wnt Pathway Effector TCF7L2 Regulates Adipocyte Development and Function". Diabetes. 67 (4): 554–568. doi: 10.2337/db17-0318. PMC 5860863. PMID 29317436.
- NCBI CDD: cd01388 (SOX-TCF_HMG-box); human proteins

The TCF/LEF family (T cell factor/lymphoid enhancer factor family) is a group of genes that encode transcription factors which bind to DNA through a SOX-like high mobility group domain. They are involved in the Wnt signaling pathway, particularly during embryonic [2] and stem-cell development, [3] but also had been found to play a role in cancer [4] and diabetes. [5] TCF/LEF factors recruit the coactivator beta-catenin to enhancer elements of genes they target. They can also recruit members of the Groucho family of corepressors. [6]
History
The discovery of the TCF/LEF genes as nuclear Wnt pathway components in the 90s [7] [8] was a pivotal breakthrough for the Wnt signalling research field, plugging an important knowledge gap and enabling subsequent understanding of transcriptional regulation of Wnt target genes, particularly in embryonic development and cancer.
Before this discovery it was only known that upstream Wnt signalling mechanisms regulated the cytoplasmic abundance of the beta-catenin protein, which as a consequence translocated into the cell nucleus. However, since the protein structure of beta-catenin did not reveal any DNA-binding domain, it was still unclear how nuclear beta-catenin could regulate Wnt target genes. Following the discovery, a model was established whereby Wnt signalling-regulated beta-catenin in the nucleus attaches to TCF/LEF DNA binding proteins, which recognise the DNA consensus sequence around the core 'CTTTG', called Wnt Response Element (WRE). [9]
This rule that beta-catenin-TCF interaction on DNA regulates Wnt target gene expression, has nonetheless been broken by examples of Wnt- and beta-catenin-independent functions for TCF/LEF proteins (for instance in zebrafish CNS development [10]) and functional association of Wnt-regulated beta-catenin with other DNA-binding transcription factors such as SOX, [11] FOXO, [12] TBX. [13] Then again, this beta-catenin-TCF interaction on DNA is now revealed as but the core of much larger protein complexes regulating transcription, called the Wnt enhanceosomes. [14] Conversely, additional mechanisms regulating TCF/LEF protein function have been discovered, such as phosphorylation [15] and sumoylation. [16]
Structure
The structure and function of TCF/LEF proteins explains this bimodal function. TCF/LEF genes encode proteins with an elaborate structure that can however be summarised by considering four main domains:
- N-terminal domain: mediating interaction with beta-catenin, which is highly conserved and mediates the transcriptional activator function.
- Control region: includes sequences regulating and mediating the transcriptional repressor function and encoding a transcriptional repressor binding domain for the Groucho family.
- DNA-binding domain: includes a very highly conserved HMG ( High Mobility Group) DNA-binding domain and the NLS ( nuclear localisation sequence).
- C-terminal tail: may contain an additional DNA-binding domain and an additional transcriptional repressor binding domain. [17]
Diversity in TCF/LEF protein structure and function comes from having different genes. Humans and jawed vertebrates generally have four genes encoding TCF/LEF proteins:
- TCF7 (also called TCF1)
- LEF1 (also called TCF1α)
- TCF7L1 (also called TCF3)
- TCF7L2 (also called TCF4)
Further diversity comes from expression from the same gene of alternative transcripts encoding different protein isoforms, particularly from the TCF7 and TCF7L2 genes:
- There are isoforms expressed from secondary promoters that encode proteins that lack the usual N-terminus and therefore specifically the beta-catenin binding domain (see above). These protein isoforms function not as bimodal transcription factors but as constitutive repressors, and they are refractory to upstream Wnt signalling regulation. [18] [19]
- There are isoforms from alternative splicing in the part of the transcript encoding the control region, which influence the propensity of the encoded protein isoforms to act as transcriptional repressors (without beta-catenin) or transcriptional activators (with beta-catenin). [20]
- There are isoforms from alternative splicing in the part of the transcript encoding the C-terminal tail resulting in protein isoforms with and without the additional DNA-binding domain and resulting in changing the reading frame in which the last exon is translated with or without an additional transcriptional co-repressor binding domain. [21] [22]
Function
TCF/LEF proteins function as bimodal transcription factors:
- As described, TCF/LEF proteins act as transcriptional activators in association with nuclear beta-catenin (and transcriptional co-activators attached to beta-catenin);
- but without beta-catenin, TCF/LEF proteins function as transcriptional repressors (attached to transcriptional co-repressors members of the Groucho family).
Thus, as a consequence, Wnt target genes are actively repressed in the absence of Wnt signalling activity, then activated when Wnt signalling actively drives beta-catenin into the nucleus. [23]
TCF/LEF genes support diverse functions in embryonic development, stem cell biology, and in disease. [24] [25] Given the conservation of structure, functions of different TCF/LEF genes and proteins are often redundant in many organs and tissues where Wnt signalling is important, yet genetic analysis suggested from the beginning that this redundancy is only partial, suggesting TCF/LEF gene- and TCF isoform-specific functions, many of which are only now beginning to be discovered.
Prominent functions of TCF/LEF genes in embryonic development include vertebrate dorsal axis induction, anterior-posterior patterning of the developing Central Nervous System, neural crest development and many functions in organ development. Prominent functions of TCF/LEF genes in stem cell development have been particularly well dissected during the hair follicle cycle. [26] [27] TCF/LEF genes have roles in many cancers, with their role in colorectal cancer possibly being the best understood. [28] However, other human diseases have also been linked to TCF/LEF genes, particularly type 2 diabetes. [29] [30]
References
- ^ Torres‐Aguila, Nuria P.; Salonna, Marika; Hoppler, Stefan; Ferrier, David E. K. (2022-02-03). "Evolutionary diversification of the canonical Wnt signaling effector TCF/LEF in chordates". Development, Growth & Differentiation. 64 (3): 120–137. doi: 10.1111/dgd.12771. hdl: 2164/18617. ISSN 0012-1592. PMC 9303524. PMID 35048372. S2CID 246161317.
- ^ Logan, Catriona Y.; Nusse, Roel (November 2004). "The WNT Signaling Pathway in Development and Disease". Annual Review of Cell and Developmental Biology. 20 (1): 781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. PMID 15473860.
- ^ Nusse, R (May 2008). "Wnt signaling and stem cell control". Cell Research. 18 (5): 523–7. doi: 10.1038/cr.2008.47. PMID 18392048. S2CID 2503910.
- ^ Zhan, T; Rindtorff, N; Boutros, M (March 2017). "Wnt signaling in cancer". Oncogene. 36 (11): 1461–1473. doi: 10.1038/onc.2016.304. PMC 5357762. PMID 27617575.
- ^ Laudes, M (April 2011). "Role of WNT signalling in the determination of human mesenchymal stem cells into preadipocytes". Journal of Molecular Endocrinology. 46 (2): R65-72. doi: 10.1530/JME-10-0169. PMID 21247979.
- ^ Brantjes H, Barker N, van Es J, Clevers H (February 2002). "TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling". Biol. Chem. 383 (2): 255–61. doi: 10.1515/BC.2002.027. PMID 11934263. S2CID 25665021.
- ^ Behrens, J; von Kries, JP; Kühl, M; Bruhn, L; Wedlich, D; Grosschedl, R; Birchmeier, W (15 August 1996). "Functional interaction of beta-catenin with the transcription factor LEF-1". Nature. 382 (6592): 638–42. Bibcode: 1996Natur.382..638B. doi: 10.1038/382638a0. PMID 8757136. S2CID 4369341.
- ^ Huber, O; Korn, R; McLaughlin, J; Ohsugi, M; Herrmann, BG; Kemler, R (September 1996). "Nuclear localization of beta-catenin by interaction with transcription factor LEF-1". Mechanisms of Development. 59 (1): 3–10. doi: 10.1016/0925-4773(96)00597-7. PMID 8892228. S2CID 14324471.
- ^ Cadigan, KM; Waterman, ML (1 November 2012). "TCF/LEFs and Wnt signaling in the nucleus". Cold Spring Harbor Perspectives in Biology. 4 (11): a007906. doi: 10.1101/cshperspect.a007906. PMC 3536346. PMID 23024173.
- ^ Duncan, RN; Panahi, S; Piotrowski, T; Dorsky, RI (2015). "Identification of Wnt Genes Expressed in Neural Progenitor Zones during Zebrafish Brain Development". PLOS ONE. 10 (12): e0145810. Bibcode: 2015PLoSO..1045810D. doi: 10.1371/journal.pone.0145810. PMC 4699909. PMID 26713625.
- ^ Kormish, JD; Sinner, D; Zorn, AM (January 2010). "Interactions between SOX factors and Wnt/beta-catenin signaling in development and disease". Developmental Dynamics. 239 (1): 56–68. doi: 10.1002/dvdy.22046. PMC 3269784. PMID 19655378.
- ^ Essers, MA; de Vries-Smits, LM; Barker, N; Polderman, PE; Burgering, BM; Korswagen, HC (20 May 2005). "Functional interaction between beta-catenin and FOXO in oxidative stress signaling". Science. 308 (5725): 1181–4. Bibcode: 2005Sci...308.1181E. doi: 10.1126/science.1109083. PMID 15905404. S2CID 24572861.
- ^ Zimmerli, Dario; Borrelli, Costanza; Jauregi-Miguel, Amaia; Söderholm, Simon; Brütsch, Salome; Doumpas, Nikolaos; Reichmuth, Jan; Murphy-Seiler, Fabienne; Aguet, MIchel; Basler, Konrad; Moor, Andreas E; Cantù, Claudio (18 August 2020). "TBX3 acts as tissue-specific component of the Wnt/β-catenin transcriptional complex". eLife. 9. doi: 10.7554/eLife.58123. PMC 7434441. PMID 32808927.
- ^ Gammons, M; Bienz, M (April 2018). "Multiprotein complexes governing Wnt signal transduction". Current Opinion in Cell Biology. 51: 42–49. doi: 10.1016/j.ceb.2017.10.008. PMID 29153704.
- ^ Sokol, SY (July 2011). "Wnt signaling through T-cell factor phosphorylation". Cell Research. 21 (7): 1002–12. doi: 10.1038/cr.2011.86. PMC 3193496. PMID 21606952.
- ^ Yamamoto, H; Ihara, M; Matsuura, Y; Kikuchi, A (1 May 2003). "Sumoylation is involved in beta-catenin-dependent activation of Tcf-4". The EMBO Journal. 22 (9): 2047–59. doi: 10.1093/emboj/cdg204. PMC 156076. PMID 12727872.
- ^ Hoppler, Stefan; Waterman, Marian L. (2014). "Evolutionary Diversification of Vertebrate TCF/LEF Structure, Function, and Regulation". WNT Signaling in Development and Disease: 225–237. doi: 10.1002/9781118444122.ch17. ISBN 9781118444122.
- ^ Van de Wetering, M; Castrop, J; Korinek, V; Clevers, H (March 1996). "Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties". Molecular and Cellular Biology. 16 (3): 745–52. doi: 10.1128/MCB.16.3.745. PMC 231054. PMID 8622675.
- ^ Hovanes, K; Li, TW; Munguia, JE; Truong, T; Milovanovic, T; Lawrence Marsh, J; Holcombe, RF; Waterman, ML (May 2001). "Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer". Nature Genetics. 28 (1): 53–7. doi: 10.1038/ng0501-53. PMID 11326276. S2CID 28974522.
- ^ Liu, F; van den Broek, O; Destrée, O; Hoppler, S (December 2005). "Distinct roles for Xenopus Tcf/Lef genes in mediating specific responses to Wnt/beta-catenin signalling in mesoderm development". Development. 132 (24): 5375–85. doi: 10.1242/dev.02152. PMID 16291789. S2CID 20515922.
- ^ Atcha, FA; Syed, A; Wu, B; Hoverter, NP; Yokoyama, NN; Ting, JH; Munguia, JE; Mangalam, HJ; Marsh, JL; Waterman, ML (December 2007). "A unique DNA binding domain converts T-cell factors into strong Wnt effectors". Molecular and Cellular Biology. 27 (23): 8352–63. doi: 10.1128/MCB.02132-06. PMC 2169181. PMID 17893322.
- ^ Ravindranath, AJ; Cadigan, KM (3 August 2016). "The Role of the C-Clamp in Wnt-Related Colorectal Cancers". Cancers. 8 (8): 74. doi: 10.3390/cancers8080074. PMC 4999783. PMID 27527215.
- ^ Ramakrishnan, AB; Cadigan, KM (2017). "Wnt target genes and where to find them". F1000Research. 6: 746. doi: 10.12688/f1000research.11034.1. PMC 5464219. PMID 28649368.
- ^ Hoppler, S; Kavanagh, CL (1 February 2007). "Wnt signalling: variety at the core". Journal of Cell Science. 120 (Pt 3): 385–93. doi: 10.1242/jcs.03363. PMID 17251379. S2CID 30976795.
-
^ Hoppler, Stefan; Moon, Randall T. (2014). Wnt signaling in development and disease : molecular mechanisms and biological functions. Hoboken, New Jersey.
ISBN
9781118444122.
{{ cite book}}: CS1 maint: location missing publisher ( link) - ^ DasGupta, R; Fuchs, E (October 1999). "Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation". Development. 126 (20): 4557–68. doi: 10.1242/dev.126.20.4557. PMID 10498690.
- ^ Merrill, BJ; Gat, U; DasGupta, R; Fuchs, E (1 July 2001). "Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin". Genes & Development. 15 (13): 1688–705. doi: 10.1101/gad.891401. PMC 312726. PMID 11445543.
- ^ Mayer, Claus-Dieter; Magon de La Giclais, Soizick; Alsehly, Fozan; Hoppler, Stefan (May 2020). "Diverse LEF/TCF Expression in Human Colorectal Cancer Correlates with Altered Wnt-Regulated Transcriptome in a Meta-Analysis of Patient Biopsies". Genes. 11 (5): 538. doi: 10.3390/genes11050538. PMC 7288467. PMID 32403323.
- ^ Jin, T; Liu, L (November 2008). "The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus". Molecular Endocrinology. 22 (11): 2383–92. doi: 10.1210/me.2008-0135. PMID 18599616.
- ^ Chen, X; Ayala, I; Shannon, C; Fourcaudot, M; Acharya, NK; Jenkinson, CP; Heikkinen, S; Norton, L (April 2018). "The Diabetes Gene and Wnt Pathway Effector TCF7L2 Regulates Adipocyte Development and Function". Diabetes. 67 (4): 554–568. doi: 10.2337/db17-0318. PMC 5860863. PMID 29317436.
- NCBI CDD: cd01388 (SOX-TCF_HMG-box); human proteins