| Mannosyl-oligosaccharide glucosidase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 3.2.1.106 | ||||||||
| CAS no. | 78413-07-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Mannosyl-oligosaccharide glucosidase (MOGS) ( EC 3.2.1.106, processing α-glucosidase I, Glc3Man9NAc2 oligosaccharide glucosidase, trimming glucosidase I, GCS1) is an enzyme with systematic name mannosyl-oligosaccharide glucohydrolase. [1] [2] [3] [4] [5] MOGS is a transmembrane protein found in the membrane of the endoplasmic reticulum of eukaryotic cells. Biologically, it functions within the N-glycosylation pathway.
Enzyme mechanism
MOGS is a glycoside hydrolase enzyme, belonging to Family 63 as classified within the Carbohydrate-Active Enzyme database. [6]
It catalyses exohydrolysis of the non-reducing terminal glucose residue in the mannosyl-oligosaccharide glycan Glc3Man9GlcNAc2.
This reaction is the first trimming step in the N-glycosylation pathway. Prior to this, the glycan was co-translationally attached to a nascent protein by the oligosaccharyltransferase complex. MOGS removes the terminal glucose residue, leaving the glycoprotein linked to Glc2Man9GlcNAc2, which can then serve as a substrate for glucosidase II.
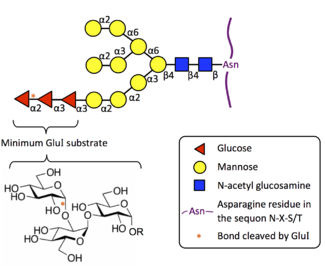
Substrate Specificity
MOGS is highly specific to the oligosaccharide in its biological substrate in the N-glycosylation pathway. Eukaryotic MOGS does not cleave simple substrates such as p-nitrophenyl glucose, and it also shows no activity to the α(1→3) linkage present at the terminus of Glc1-2Man9GlcNAc2. [7] [8] [9] Furthermore, the minimum substrate is the glucotriose molecule (Glc-α(1→2)-Glc-α(1→3)-Glc), linked as in its native Glc3Man9GlcNAc2 substrate. Kojibiose, the disaccharide Glc-α(1→2)-Glc, acts as a weak inhibitor on plant, animal, and yeast MOGS. [8] [10] [11] [12]
MOGS also acts to lesser extent on the corresponding glycolipids and glycopeptides.
References
- ^ Elting JJ, Chen WW, Lennarz WJ (March 1980). "Characterization of a glucosidase involved in an initial step in the processing of oligosaccharide chains". The Journal of Biological Chemistry. 255 (6): 2325–31. PMID 7358674.
- ^ Grinna LS, Robbins PW (September 1979). "Glycoprotein biosynthesis. Rat liver microsomal glucosidases which process oligosaccharides". The Journal of Biological Chemistry. 254 (18): 8814–8. PMID 479161.
- ^ Kilker RD, Saunier B, Tkacz JS, Herscovics A (May 1981). "Partial purification from Saccharomyces cerevisiae of a soluble glucosidase which removes the terminal glucose from the oligosaccharide Glc3Man9GlcNAc2". The Journal of Biological Chemistry. 256 (10): 5299–603. PMID 7014569.
- ^ Grinna LS, Robbins PW (March 1980). "Substrate specificities of rat liver microsomal glucosidases which process glycoproteins". The Journal of Biological Chemistry. 255 (6): 2255–8. PMID 7358666.
- ^ Michael JM, Kornfeld S (January 1980). "Partial purification and characterization of the glucosidases involved in the processing of asparagine-linked oligosaccharides". Archives of Biochemistry and Biophysics. 199 (1): 249–58. doi: 10.1016/0003-9861(80)90278-7. PMID 7356331.
- ^ "CAZy - GH63". www.cazy.org. Retrieved 2016-04-05.
- ^ Vijay IK, Shailubhai K, Dong-Yu B, Pratta MA, Saxena S (1988-04-01). "Studies on the biosynthesis and regulation of asparagine-linked glycoproteins in the lactating mammary gland". Indian Journal of Biochemistry & Biophysics. 25 (1–2): 127–32. PMID 2846425.
- ^ a b Dhanawansa R, Faridmoayer A, van der Merwe G, Li YX, Scaman CH (March 2002). "Overexpression, purification, and partial characterization of Saccharomyces cerevisiae processing α glucosidase I". Glycobiology. 12 (3): 229–34. doi: 10.1016/0014-5793(86)80982-6. PMID 11971867.
- ^ Shailubhai K, Saxena ES, Balapure AK, Vijay IK (June 1990). "Developmental regulation of glucosidase I, an enzyme involved in the processing of asparagine-linked glycoproteins in rat mammary gland". The Journal of Biological Chemistry. 265 (17): 9701–6. PMID 2190984.
- ^ Zeng YC, Elbein AD (July 1998). "Purification to homogeneity and properties of plant glucosidase I". Archives of Biochemistry and Biophysics. 355 (1): 26–34. doi: 10.1006/abbi.1998.0717. PMID 9647663.
- ^ Schweden J, Borgmann C, Legler G, Bause E (July 1986). "Characterization of calf liver glucosidase I and its inhibition by basic sugar analogs". Archives of Biochemistry and Biophysics. 248 (1): 335–40. doi: 10.1016/0003-9861(86)90429-7. PMID 2942110.
- ^ Ugalde RA, Staneloni RJ, Leloir LF (December 1980). "Microsomal glucosidases of rat liver. Partial purification and inhibition by disaccharides". European Journal of Biochemistry. 113 (1): 97–103. doi: 10.1111/j.1432-1033.1980.tb06144.x. hdl: 11336/143170. PMID 7460954.
External links
- Mannosyl-oligosaccharide+glucosidase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
| Mannosyl-oligosaccharide glucosidase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 3.2.1.106 | ||||||||
| CAS no. | 78413-07-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Mannosyl-oligosaccharide glucosidase (MOGS) ( EC 3.2.1.106, processing α-glucosidase I, Glc3Man9NAc2 oligosaccharide glucosidase, trimming glucosidase I, GCS1) is an enzyme with systematic name mannosyl-oligosaccharide glucohydrolase. [1] [2] [3] [4] [5] MOGS is a transmembrane protein found in the membrane of the endoplasmic reticulum of eukaryotic cells. Biologically, it functions within the N-glycosylation pathway.
Enzyme mechanism
MOGS is a glycoside hydrolase enzyme, belonging to Family 63 as classified within the Carbohydrate-Active Enzyme database. [6]
It catalyses exohydrolysis of the non-reducing terminal glucose residue in the mannosyl-oligosaccharide glycan Glc3Man9GlcNAc2.
This reaction is the first trimming step in the N-glycosylation pathway. Prior to this, the glycan was co-translationally attached to a nascent protein by the oligosaccharyltransferase complex. MOGS removes the terminal glucose residue, leaving the glycoprotein linked to Glc2Man9GlcNAc2, which can then serve as a substrate for glucosidase II.

Substrate Specificity
MOGS is highly specific to the oligosaccharide in its biological substrate in the N-glycosylation pathway. Eukaryotic MOGS does not cleave simple substrates such as p-nitrophenyl glucose, and it also shows no activity to the α(1→3) linkage present at the terminus of Glc1-2Man9GlcNAc2. [7] [8] [9] Furthermore, the minimum substrate is the glucotriose molecule (Glc-α(1→2)-Glc-α(1→3)-Glc), linked as in its native Glc3Man9GlcNAc2 substrate. Kojibiose, the disaccharide Glc-α(1→2)-Glc, acts as a weak inhibitor on plant, animal, and yeast MOGS. [8] [10] [11] [12]
MOGS also acts to lesser extent on the corresponding glycolipids and glycopeptides.
References
- ^ Elting JJ, Chen WW, Lennarz WJ (March 1980). "Characterization of a glucosidase involved in an initial step in the processing of oligosaccharide chains". The Journal of Biological Chemistry. 255 (6): 2325–31. PMID 7358674.
- ^ Grinna LS, Robbins PW (September 1979). "Glycoprotein biosynthesis. Rat liver microsomal glucosidases which process oligosaccharides". The Journal of Biological Chemistry. 254 (18): 8814–8. PMID 479161.
- ^ Kilker RD, Saunier B, Tkacz JS, Herscovics A (May 1981). "Partial purification from Saccharomyces cerevisiae of a soluble glucosidase which removes the terminal glucose from the oligosaccharide Glc3Man9GlcNAc2". The Journal of Biological Chemistry. 256 (10): 5299–603. PMID 7014569.
- ^ Grinna LS, Robbins PW (March 1980). "Substrate specificities of rat liver microsomal glucosidases which process glycoproteins". The Journal of Biological Chemistry. 255 (6): 2255–8. PMID 7358666.
- ^ Michael JM, Kornfeld S (January 1980). "Partial purification and characterization of the glucosidases involved in the processing of asparagine-linked oligosaccharides". Archives of Biochemistry and Biophysics. 199 (1): 249–58. doi: 10.1016/0003-9861(80)90278-7. PMID 7356331.
- ^ "CAZy - GH63". www.cazy.org. Retrieved 2016-04-05.
- ^ Vijay IK, Shailubhai K, Dong-Yu B, Pratta MA, Saxena S (1988-04-01). "Studies on the biosynthesis and regulation of asparagine-linked glycoproteins in the lactating mammary gland". Indian Journal of Biochemistry & Biophysics. 25 (1–2): 127–32. PMID 2846425.
- ^ a b Dhanawansa R, Faridmoayer A, van der Merwe G, Li YX, Scaman CH (March 2002). "Overexpression, purification, and partial characterization of Saccharomyces cerevisiae processing α glucosidase I". Glycobiology. 12 (3): 229–34. doi: 10.1016/0014-5793(86)80982-6. PMID 11971867.
- ^ Shailubhai K, Saxena ES, Balapure AK, Vijay IK (June 1990). "Developmental regulation of glucosidase I, an enzyme involved in the processing of asparagine-linked glycoproteins in rat mammary gland". The Journal of Biological Chemistry. 265 (17): 9701–6. PMID 2190984.
- ^ Zeng YC, Elbein AD (July 1998). "Purification to homogeneity and properties of plant glucosidase I". Archives of Biochemistry and Biophysics. 355 (1): 26–34. doi: 10.1006/abbi.1998.0717. PMID 9647663.
- ^ Schweden J, Borgmann C, Legler G, Bause E (July 1986). "Characterization of calf liver glucosidase I and its inhibition by basic sugar analogs". Archives of Biochemistry and Biophysics. 248 (1): 335–40. doi: 10.1016/0003-9861(86)90429-7. PMID 2942110.
- ^ Ugalde RA, Staneloni RJ, Leloir LF (December 1980). "Microsomal glucosidases of rat liver. Partial purification and inhibition by disaccharides". European Journal of Biochemistry. 113 (1): 97–103. doi: 10.1111/j.1432-1033.1980.tb06144.x. hdl: 11336/143170. PMID 7460954.
External links
- Mannosyl-oligosaccharide+glucosidase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)