| Rhodanese-like domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| Symbol | Rhodanese | ||||||||
| Pfam | PF00581 | ||||||||
| InterPro | IPR001763 | ||||||||
| PROSITE | PDOC00322 | ||||||||
| SCOP2 | 2ora / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 413 | ||||||||
| OPM protein | 2mpn | ||||||||
| CDD | cd00158 | ||||||||
| Membranome | 571 | ||||||||
| |||||||||
| thiosulfate sulfurtransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 2.8.1.1 | ||||||||
| CAS no. | 9026-04-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Rhodanese is a mitochondrial enzyme that detoxifies cyanide (CN−) by converting it to thiocyanate (SCN−, also known as "rhodanate"). [1] In enzymatology, the common name is listed as thiosulfate sulfurtransferase ( EC 2.8.1.1). [2] The diagram on the right shows the crystallographically-determined structure of rhodanese.
It catalyzes the following reaction:
- thiosulfate + cyanide sulfite + thiocyanate
Structure and mechanism
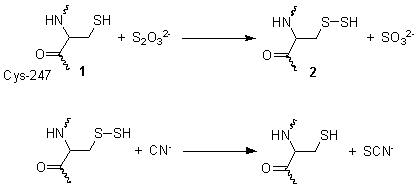
This reaction takes place in two steps. In the first step, thiosulfate is reduced by the thiol group on cysteine-247 1, to form a persulfide and a sulfite 2. In the second step, the persulfide reacts with cyanide to produce thiocyanate, re-generating the cysteine thiol 1. [3]
Rhodanese shares evolutionary relationship with a large family of proteins, including:
- Cdc25 phosphatase catalytic domain
- non-catalytic domains of eukaryotic dual-specificity MAPK-phosphatases
- non-catalytic domains of yeast PTP-type MAPK-phosphatases
- non-catalytic domains of yeast Ubp4, Ubp5, Ubp7
- non-catalytic domains of mammalian Ubp-Y
- Drosophila heat shock protein HSP-67BB
- several bacterial cold-shock and phage shock proteins
- plant senescence associated proteins
- catalytic and non-catalytic domains of rhodanese [4]
Rhodanese has an internal duplication. This domain is found as a single copy in other proteins, including phosphatases and ubiquitin C-terminal hydrolases. [5]
Clinical relevance
This reaction is important for the treatment of exposure to cyanide, since the thiocyanate formed is around 1 / 200 as toxic. [6]:p. 15938 The use of thiosulfate solution as an antidote for cyanide poisoning is based on the activation of this enzymatic cycle.
Human proteins
The human mitochondrial rhodanese gene is TST.
The following other human genes match the "Rhodanese-like" domain on InterPro, but are not the rodanase with its catalytic activity (see also the list of related families in #Structure and mechanism):
- M-phase inducer phosphatase: CDC25A; CDC25B; CDC25C;
- Dual specificity protein phosphatase: DUSP; DUSP1; DUSP2; DUSP4; DUSP5; DUSP6; DUSP7; DUSP10; DUSP16, aka MKP7;
- Thiosulfate:glutathione sulfurtransferase: KAT, now known as " TSTD1";
- Adenylyltransferase and sulfurtransferase: MOCS3;
- 3-mercaptopyruvate sulfurtransferase: MPST, also known as "TSTD2"
- Not an enzyme: TBCK; TSGA14;
- Ubiquitin carboxyl-terminal hydrolase: USP8;
- Unknown activity: TSTD3
Nomenclature
Although the standard nomenclature rules for enzymes indicate that their names are to end with the letters "-ase", rhodanese was first described in 1933, [7] prior to the 1955 establishment of the Enzyme Commission; as such, the older name had already attained widespread usage.
The systematic name of this enzyme class is "thiosulfate:cyanide sulfurtransferase". Other names in common use include "thiosulfate cyanide transsulfurase", "thiosulfate thiotransferase", "rhodanese", and "rhodanase".
References
- ^ Cipollone R, Ascenzi P, Tomao P, Imperi F, Visca P (2008). "Enzymatic detoxification of cyanide: clues from Pseudomonas aeruginosa Rhodanese". Journal of Molecular Microbiology and Biotechnology. 15 (2–3): 199–211. doi: 10.1159/000121331. PMID 18685272. S2CID 25431686.
- ^ EC 2.8.1.1, at the International Union of Biochemistry and Molecular Biology
- ^ Cipollone R, Ascenzi P, Tomao P, Imperi F, Visca P (2008). "Enzymatic detoxification of cyanide: clues from Pseudomonas aeruginosa Rhodanese". Journal of Molecular Microbiology and Biotechnology. 15 (2–3): 199–211. doi: 10.1159/000121331. PMID 18685272. S2CID 25431686.
- ^ "Thiosulphate sulfurtransferase, conserved site (IPR001307)". EMBL-EBI. InterPro.
- ^ Gliubich F, Gazerro M, Zanotti G, Delbono S, Bombieri G, Berni R (August 1996). "Active site structural features for chemically modified forms of rhodanese". The Journal of Biological Chemistry. 271 (35): 21054–61. doi: 10.1074/jbc.271.35.21054. PMID 8702871.
- ^ Jaszczak E, Polkowska Ż, Narkowicz S, Namieśnik J (July 2017). "Cyanides in the environment-analysis-problems and challenges". Environmental Science and Pollution Research International. 24 (19): 15929–15948. doi: 10.1007/s11356-017-9081-7. PMC 5506515. PMID 28512706.
- ^ Cipollone R, Ascenzi P, Visca P (February 2007). "Common themes and variations in the rhodanese superfamily". IUBMB Life. 59 (2): 51–9. doi: 10.1080/15216540701206859. PMID 17454295.
External links
- Rhodanese at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
| Rhodanese-like domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| Symbol | Rhodanese | ||||||||
| Pfam | PF00581 | ||||||||
| InterPro | IPR001763 | ||||||||
| PROSITE | PDOC00322 | ||||||||
| SCOP2 | 2ora / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 413 | ||||||||
| OPM protein | 2mpn | ||||||||
| CDD | cd00158 | ||||||||
| Membranome | 571 | ||||||||
| |||||||||
| thiosulfate sulfurtransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 2.8.1.1 | ||||||||
| CAS no. | 9026-04-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Rhodanese is a mitochondrial enzyme that detoxifies cyanide (CN−) by converting it to thiocyanate (SCN−, also known as "rhodanate"). [1] In enzymatology, the common name is listed as thiosulfate sulfurtransferase ( EC 2.8.1.1). [2] The diagram on the right shows the crystallographically-determined structure of rhodanese.
It catalyzes the following reaction:
- thiosulfate + cyanide sulfite + thiocyanate
Structure and mechanism
This reaction takes place in two steps. In the first step, thiosulfate is reduced by the thiol group on cysteine-247 1, to form a persulfide and a sulfite 2. In the second step, the persulfide reacts with cyanide to produce thiocyanate, re-generating the cysteine thiol 1. [3]
Rhodanese shares evolutionary relationship with a large family of proteins, including:
- Cdc25 phosphatase catalytic domain
- non-catalytic domains of eukaryotic dual-specificity MAPK-phosphatases
- non-catalytic domains of yeast PTP-type MAPK-phosphatases
- non-catalytic domains of yeast Ubp4, Ubp5, Ubp7
- non-catalytic domains of mammalian Ubp-Y
- Drosophila heat shock protein HSP-67BB
- several bacterial cold-shock and phage shock proteins
- plant senescence associated proteins
- catalytic and non-catalytic domains of rhodanese [4]
Rhodanese has an internal duplication. This domain is found as a single copy in other proteins, including phosphatases and ubiquitin C-terminal hydrolases. [5]
Clinical relevance
This reaction is important for the treatment of exposure to cyanide, since the thiocyanate formed is around 1 / 200 as toxic. [6]:p. 15938 The use of thiosulfate solution as an antidote for cyanide poisoning is based on the activation of this enzymatic cycle.
Human proteins
The human mitochondrial rhodanese gene is TST.
The following other human genes match the "Rhodanese-like" domain on InterPro, but are not the rodanase with its catalytic activity (see also the list of related families in #Structure and mechanism):
- M-phase inducer phosphatase: CDC25A; CDC25B; CDC25C;
- Dual specificity protein phosphatase: DUSP; DUSP1; DUSP2; DUSP4; DUSP5; DUSP6; DUSP7; DUSP10; DUSP16, aka MKP7;
- Thiosulfate:glutathione sulfurtransferase: KAT, now known as " TSTD1";
- Adenylyltransferase and sulfurtransferase: MOCS3;
- 3-mercaptopyruvate sulfurtransferase: MPST, also known as "TSTD2"
- Not an enzyme: TBCK; TSGA14;
- Ubiquitin carboxyl-terminal hydrolase: USP8;
- Unknown activity: TSTD3
Nomenclature
Although the standard nomenclature rules for enzymes indicate that their names are to end with the letters "-ase", rhodanese was first described in 1933, [7] prior to the 1955 establishment of the Enzyme Commission; as such, the older name had already attained widespread usage.
The systematic name of this enzyme class is "thiosulfate:cyanide sulfurtransferase". Other names in common use include "thiosulfate cyanide transsulfurase", "thiosulfate thiotransferase", "rhodanese", and "rhodanase".
References
- ^ Cipollone R, Ascenzi P, Tomao P, Imperi F, Visca P (2008). "Enzymatic detoxification of cyanide: clues from Pseudomonas aeruginosa Rhodanese". Journal of Molecular Microbiology and Biotechnology. 15 (2–3): 199–211. doi: 10.1159/000121331. PMID 18685272. S2CID 25431686.
- ^ EC 2.8.1.1, at the International Union of Biochemistry and Molecular Biology
- ^ Cipollone R, Ascenzi P, Tomao P, Imperi F, Visca P (2008). "Enzymatic detoxification of cyanide: clues from Pseudomonas aeruginosa Rhodanese". Journal of Molecular Microbiology and Biotechnology. 15 (2–3): 199–211. doi: 10.1159/000121331. PMID 18685272. S2CID 25431686.
- ^ "Thiosulphate sulfurtransferase, conserved site (IPR001307)". EMBL-EBI. InterPro.
- ^ Gliubich F, Gazerro M, Zanotti G, Delbono S, Bombieri G, Berni R (August 1996). "Active site structural features for chemically modified forms of rhodanese". The Journal of Biological Chemistry. 271 (35): 21054–61. doi: 10.1074/jbc.271.35.21054. PMID 8702871.
- ^ Jaszczak E, Polkowska Ż, Narkowicz S, Namieśnik J (July 2017). "Cyanides in the environment-analysis-problems and challenges". Environmental Science and Pollution Research International. 24 (19): 15929–15948. doi: 10.1007/s11356-017-9081-7. PMC 5506515. PMID 28512706.
- ^ Cipollone R, Ascenzi P, Visca P (February 2007). "Common themes and variations in the rhodanese superfamily". IUBMB Life. 59 (2): 51–9. doi: 10.1080/15216540701206859. PMID 17454295.
External links
- Rhodanese at the U.S. National Library of Medicine Medical Subject Headings (MeSH)