| Cytochrome c nitrite reductase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
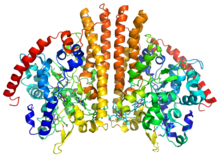
Crystallographic structure of a
homodimer of the cytochrome c nitrite reductase from
Escherichia coli (rainbow colored cartoon, blue =
N-terminus, red =
C-terminus) complexed with
heme C (sticks).
[1] | |||||||||
| Identifiers | |||||||||
| EC no. | 1.7.2.2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Cytochrome c nitrite reductase (ccNiR) ( EC 1.7.2.2) is a bacterial enzyme that catalyzes the six electron reduction of nitrite to ammonia; an important step in the biological nitrogen cycle. [2] The enzyme catalyses the second step in the two step conversion of nitrate to ammonia, which allows certain bacteria to use nitrite as a terminal electron acceptor, rather than oxygen, during anaerobic conditions. During this process, ccNiR draws electrons from the quinol pool, which are ultimately provided by a dehydrogenase such as formate dehydrogenase or hydrogenase. These dehydrogenases are responsible for generating a proton motive force. [3]
Cytochrome c Nitrite Reductase is a homodimer which contains five c-type heme cofactors per monomer. [4] Four of the heme centers are bis-histidine ligated and presumably serve to shuttle electrons to the active site. The active site heme, however, is uniquely ligated by a single lysine residue.
This enzyme belongs to the family of oxidoreductases, specifically those acting on other nitrogenous compounds as donors with a cytochrome as acceptor. The systematic name of this enzyme class is ammonia:ferricytochrome-c oxidoreductase.
| Cytochrom_NNT | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Cytochrom_NNT | ||||||||
| Pfam | PF03264 | ||||||||
| Pfam clan | CL0317 | ||||||||
| InterPro | IPR005126 | ||||||||
| OPM superfamily | 175 | ||||||||
| OPM protein | 2j7a | ||||||||
| |||||||||
References
- ^ PDB: 1GU6; Bamford VA, Angove HC, Seward HE, Thomson AJ, Cole JA, Butt JN, Hemmings AM, Richardson DJ (March 2002). "Structure and spectroscopy of the periplasmic cytochrome c nitrite reductase from Escherichia coli". Biochemistry. 41 (9): 2921–31. doi: 10.1021/bi015765d. PMID 11863430.
- ^ Clarke TA, Mills PC, Poock SR, Butt JN, Cheesman MR, Cole JA, Hinton JC, Hemmings AM, Kemp G, Söderberg CA, Spiro S, Van Wonderen J, Richardson DJ (2008). "Escherichia coli Cytochrome c Nitrite Reductase NrfA". Globins and Other Nitric Oxide-Reactive Proteins, Part B. Methods in Enzymology. Vol. 437. pp. 63–77. doi: 10.1016/S0076-6879(07)37004-3. ISBN 9780123742780. PMID 18433623.
- ^ Simon J (August 2002). "Enzymology and bioenergetics of respiratory nitrite ammonification". FEMS Microbiol. Rev. 26 (3): 285–309. doi: 10.1111/j.1574-6976.2002.tb00616.x. PMID 12165429.
- ^ Einsle O, Messerschmidt A, Stach P, Bourenkov GP, Bartunik HD, Huber R, Kroneck PM (July 1999). "Structure of cytochrome c nitrite reductase". Nature. 400 (6743): 476–80. Bibcode: 1999Natur.400..476E. doi: 10.1038/22802. PMID 10440380. S2CID 4403452.
Further reading
- Einsle O, Messerschmidt A, Stach P, Bourenkov GP, Bartunik HD, Huber R, Kroneck PM (July 1999). "Structure of cytochrome c nitrite reductase". Nature. 400 (6743): 476–80. Bibcode: 1999Natur.400..476E. doi: 10.1038/22802. PMID 10440380. S2CID 4403452.
| Cytochrome c nitrite reductase | |||||||||
|---|---|---|---|---|---|---|---|---|---|

Crystallographic structure of a
homodimer of the cytochrome c nitrite reductase from
Escherichia coli (rainbow colored cartoon, blue =
N-terminus, red =
C-terminus) complexed with
heme C (sticks).
[1] | |||||||||
| Identifiers | |||||||||
| EC no. | 1.7.2.2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Cytochrome c nitrite reductase (ccNiR) ( EC 1.7.2.2) is a bacterial enzyme that catalyzes the six electron reduction of nitrite to ammonia; an important step in the biological nitrogen cycle. [2] The enzyme catalyses the second step in the two step conversion of nitrate to ammonia, which allows certain bacteria to use nitrite as a terminal electron acceptor, rather than oxygen, during anaerobic conditions. During this process, ccNiR draws electrons from the quinol pool, which are ultimately provided by a dehydrogenase such as formate dehydrogenase or hydrogenase. These dehydrogenases are responsible for generating a proton motive force. [3]
Cytochrome c Nitrite Reductase is a homodimer which contains five c-type heme cofactors per monomer. [4] Four of the heme centers are bis-histidine ligated and presumably serve to shuttle electrons to the active site. The active site heme, however, is uniquely ligated by a single lysine residue.
This enzyme belongs to the family of oxidoreductases, specifically those acting on other nitrogenous compounds as donors with a cytochrome as acceptor. The systematic name of this enzyme class is ammonia:ferricytochrome-c oxidoreductase.
| Cytochrom_NNT | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Cytochrom_NNT | ||||||||
| Pfam | PF03264 | ||||||||
| Pfam clan | CL0317 | ||||||||
| InterPro | IPR005126 | ||||||||
| OPM superfamily | 175 | ||||||||
| OPM protein | 2j7a | ||||||||
| |||||||||
References
- ^ PDB: 1GU6; Bamford VA, Angove HC, Seward HE, Thomson AJ, Cole JA, Butt JN, Hemmings AM, Richardson DJ (March 2002). "Structure and spectroscopy of the periplasmic cytochrome c nitrite reductase from Escherichia coli". Biochemistry. 41 (9): 2921–31. doi: 10.1021/bi015765d. PMID 11863430.
- ^ Clarke TA, Mills PC, Poock SR, Butt JN, Cheesman MR, Cole JA, Hinton JC, Hemmings AM, Kemp G, Söderberg CA, Spiro S, Van Wonderen J, Richardson DJ (2008). "Escherichia coli Cytochrome c Nitrite Reductase NrfA". Globins and Other Nitric Oxide-Reactive Proteins, Part B. Methods in Enzymology. Vol. 437. pp. 63–77. doi: 10.1016/S0076-6879(07)37004-3. ISBN 9780123742780. PMID 18433623.
- ^ Simon J (August 2002). "Enzymology and bioenergetics of respiratory nitrite ammonification". FEMS Microbiol. Rev. 26 (3): 285–309. doi: 10.1111/j.1574-6976.2002.tb00616.x. PMID 12165429.
- ^ Einsle O, Messerschmidt A, Stach P, Bourenkov GP, Bartunik HD, Huber R, Kroneck PM (July 1999). "Structure of cytochrome c nitrite reductase". Nature. 400 (6743): 476–80. Bibcode: 1999Natur.400..476E. doi: 10.1038/22802. PMID 10440380. S2CID 4403452.
Further reading
- Einsle O, Messerschmidt A, Stach P, Bourenkov GP, Bartunik HD, Huber R, Kroneck PM (July 1999). "Structure of cytochrome c nitrite reductase". Nature. 400 (6743): 476–80. Bibcode: 1999Natur.400..476E. doi: 10.1038/22802. PMID 10440380. S2CID 4403452.