This article may be too technical for most readers to understand. (August 2016) |
| Vts1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystal structure of Vts1p–SRE complex.
[1] | |||||||||
| Identifiers | |||||||||
| Symbol | Vts1 | ||||||||
| Pfam | PF07647 | ||||||||
| SCOP2 | 1b0x / SCOPe / SUPFAM | ||||||||
| |||||||||
Vts1 is a post-transcriptional regulator that has RNA-binding Sterile alpha motif (SAM) domain. [2] [3] The protein is found in Saccharomyces cerevisiae and several eukaryotes. In Saccharomyces the Vts1 impacts vesicular transport and sporulation. [4] [5]
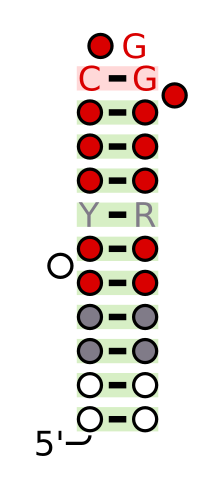
Protein-protein interactions through SAM domains participate in different regulatory activities such as signal transduction. Proteins having such domains were also shown to recognize and interact with RNA structures of similar shape to the Smaug response element (SRE). [6] Vts1 binds to RNA targets that have CUGGC on hairpin loops. [7]
- ^ Johnson PE, Donaldson LW (February 2006). "RNA recognition by the Vts1p SAM domain". Nature Structural & Molecular Biology. 13 (2): 177–178. doi: 10.1038/nsmb1039. PMID 16429155. S2CID 9561521.
- ^ Schultz J, Ponting CP, Hofmann K, Bork P (January 1997). "SAM as a protein interaction domain involved in developmental regulation". Protein Science. 6 (1): 249–253. doi: 10.1002/pro.5560060128. PMC 2143507. PMID 9007998.
- ^ Oberstrass FC, Lee A, Stefl R, Janis M, Chanfreau G, Allain FH (February 2006). "Shape-specific recognition in the structure of the Vts1p SAM domain with RNA". Nature Structural & Molecular Biology. 13 (2): 160–167. doi: 10.1038/nsmb1038. PMID 16429156. S2CID 15416866.
- ^ Deutschbauer AM, Williams RM, Chu AM, Davis RW (November 2002). "Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae". Proceedings of the National Academy of Sciences of the United States of America. 99 (24): 15530–15535. Bibcode: 2002PNAS...9915530D. doi: 10.1073/pnas.202604399. PMC 137751. PMID 12432101.
- ^ Dilcher M, Köhler B, von Mollard GF (September 2001). "Genetic interactions with the yeast Q-SNARE VTI1 reveal novel functions for the R-SNARE YKT6". The Journal of Biological Chemistry. 276 (37): 34537–34544. doi: 10.1074/jbc.M101551200. PMID 11445562.
- ^ Hall TM (September 2003). "SAM breaks its stereotype". Nature Structural Biology. 10 (9): 677–679. doi: 10.1038/nsb0903-677. PMID 12942139. S2CID 9072100.
- ^ Oberstrass FC, Lee A, Stefl R, Janis M, Chanfreau G, Allain FH (February 2006). "Shape-specific recognition in the structure of the Vts1p SAM domain with RNA". Nature Structural & Molecular Biology. 13 (2): 160–167. doi: 10.1038/nsmb1038. PMID 16429156. S2CID 15416866.
This article may be too technical for most readers to understand. (August 2016) |
| Vts1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystal structure of Vts1p–SRE complex.
[1] | |||||||||
| Identifiers | |||||||||
| Symbol | Vts1 | ||||||||
| Pfam | PF07647 | ||||||||
| SCOP2 | 1b0x / SCOPe / SUPFAM | ||||||||
| |||||||||
Vts1 is a post-transcriptional regulator that has RNA-binding Sterile alpha motif (SAM) domain. [2] [3] The protein is found in Saccharomyces cerevisiae and several eukaryotes. In Saccharomyces the Vts1 impacts vesicular transport and sporulation. [4] [5]

Protein-protein interactions through SAM domains participate in different regulatory activities such as signal transduction. Proteins having such domains were also shown to recognize and interact with RNA structures of similar shape to the Smaug response element (SRE). [6] Vts1 binds to RNA targets that have CUGGC on hairpin loops. [7]
- ^ Johnson PE, Donaldson LW (February 2006). "RNA recognition by the Vts1p SAM domain". Nature Structural & Molecular Biology. 13 (2): 177–178. doi: 10.1038/nsmb1039. PMID 16429155. S2CID 9561521.
- ^ Schultz J, Ponting CP, Hofmann K, Bork P (January 1997). "SAM as a protein interaction domain involved in developmental regulation". Protein Science. 6 (1): 249–253. doi: 10.1002/pro.5560060128. PMC 2143507. PMID 9007998.
- ^ Oberstrass FC, Lee A, Stefl R, Janis M, Chanfreau G, Allain FH (February 2006). "Shape-specific recognition in the structure of the Vts1p SAM domain with RNA". Nature Structural & Molecular Biology. 13 (2): 160–167. doi: 10.1038/nsmb1038. PMID 16429156. S2CID 15416866.
- ^ Deutschbauer AM, Williams RM, Chu AM, Davis RW (November 2002). "Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae". Proceedings of the National Academy of Sciences of the United States of America. 99 (24): 15530–15535. Bibcode: 2002PNAS...9915530D. doi: 10.1073/pnas.202604399. PMC 137751. PMID 12432101.
- ^ Dilcher M, Köhler B, von Mollard GF (September 2001). "Genetic interactions with the yeast Q-SNARE VTI1 reveal novel functions for the R-SNARE YKT6". The Journal of Biological Chemistry. 276 (37): 34537–34544. doi: 10.1074/jbc.M101551200. PMID 11445562.
- ^ Hall TM (September 2003). "SAM breaks its stereotype". Nature Structural Biology. 10 (9): 677–679. doi: 10.1038/nsb0903-677. PMID 12942139. S2CID 9072100.
- ^ Oberstrass FC, Lee A, Stefl R, Janis M, Chanfreau G, Allain FH (February 2006). "Shape-specific recognition in the structure of the Vts1p SAM domain with RNA". Nature Structural & Molecular Biology. 13 (2): 160–167. doi: 10.1038/nsmb1038. PMID 16429156. S2CID 15416866.