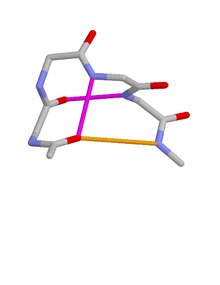
Schellman loops (also called Schellman motifs or paperclips) [1] [2] [3] [4] [5] [6] [7] [8] [9] [10] are commonly occurring structural features of proteins and polypeptides. [11] Each has six amino acid residues (labelled residues i to i+5) with two specific inter-mainchain hydrogen bonds (as in lower figure, i) and a characteristic main chain dihedral angle conformation. The CO group of residue i is hydrogen-bonded to the NH of residue i+5 (colored orange in upper figure), and the CO group of residue i+1 is hydrogen-bonded to the NH of residue i+4 ( beta turn, colored purple). Residues i+1, i+2, and i+3 have negative φ (phi) angle values and the phi value of residue i+4 is positive. Schellman loops incorporate a three amino acid residue RL nest (protein structural motif), [12] [13] in which three mainchain NH groups (from Schellman loop residues i+3 to i+5) form a concavity for hydrogen bonding to carbonyl oxygens. About 2.5% of amino acids in proteins belong to Schellman loops. Two websites are available for examining small motifs in proteins, Motivated Proteins: [1]; [14] or PDBeMotif: [2]. [15]
The majority of Schellman loops (82%) occur at the C-terminus of an alpha-helix such that residues i, i+1, i+2 and i+3 are part of the helix. Over a quarter of helices (28%) have a C-terminal Schellman loop. [10]
Occasional Schellman loops occur with seven instead of six residues. In these, the CO group of residue i is hydrogen-bonded to the NH of residue i+6, and the CO group of residue i+1 is hydrogen-bonded to the NH of residue i+5. Rare “left-handed” six-residue Schellman loops occur; these have the same hydrogen bonds, but residues i+1, i+2, and i+3 have positive φ values while the φ value of residue i+4 is negative; the nest is of the LR, rather than the RL, kind.
Amino acid propensities for the residues of the common type of Schellman loop have been described. [16] Residue i+4 is the one most-highly conserved; it has positive φ values; 70% of amino acids are glycine and none are proline.
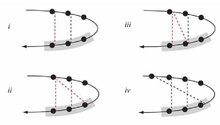
Consideration of the hydrogen bonding in the nests of Schellman loops bound to mainchain oxygens reveals two main types of arrangement: 1,3-bridged or not. In one (lower figure, ii) the first and third nest NH groups are bridged by an oxygen atom. In the other (lower figure, iv) the first NH group is hydrogen bonded to the CO group of an amino acid four residues behind in the sequence, and none of the nest NH groups are bridged. [17] It seems that Schellman loops are less homogeneous than might have been expected.
The original Schellman criteria [1] result in the inclusion of features not now regarded as Schellman loops. A newer set of criteria is given in the first paragraph.
References
- ^ a b Schellman, C (1980). Protein Folding. Amsterdam: Elsevier. pp. 53–61.
- ^ Milner-White, E J (1988-02-05). "Recurring loop motif in proteins that occurs in right-handed and left-handed forms. Its relationship with alpha-helices and beta-bulge loops". Journal of Molecular Biology. 199 (3): 503–511. doi: 10.1016/0022-2836(88)90621-3. ISSN 0022-2836. PMID 3351939.
- ^ Aurora, R; Srinivasan, R; Rose, G D (1994). "Rules for alpha-helix termination by glycine" (PDF). Science. 264 (5162): 1126–1130. doi: 10.1126/science.8178170. ISSN 0036-8075. PMID 8178170.
- ^ Viguera, Ana Rosa; Serrano, Luis (1995-08-04). "Experimental Analysis of the Schellman Motif". Journal of Molecular Biology. 251 (1): 150–160. doi: 10.1006/jmbi.1995.0422. ISSN 0022-2836. PMID 7643384.
- ^ Aurora, R; Rose, G D (January 1998). "Helix capping". Protein Science. 7 (1): 21–38. doi: 10.1002/pro.5560070103. ISSN 0961-8368. PMC 2143812. PMID 9514257.
- ^ Kallenbach, Neville R.; Gong, Youxiang (January 1999). "C-Terminal capping motifs in model helical peptides". Bioorganic & Medicinal Chemistry. 7 (1): 143–151. doi: 10.1016/S0968-0896(98)00231-4. ISSN 0968-0896. PMID 10199664.
- ^ Sukumar, Muppalla; Gierasch, Lila M (August 1997). "Local interactions in a Schellman motif dictate interhelical arrangement in a protein fragment". Folding & Design. 2 (4): 211–222. doi: 10.1016/S1359-0278(97)00030-8. ISSN 1359-0278. PMID 9269562.
- ^ Datta, Saumen; Uma, Manjappara V.; Shamala, N.; Balaram, P. (1999). "Stereochemistry of Schellman motifs in peptides: crystal structure of a hexapeptide with a C-terminus 6→1 hydrogen bond" (PDF). Biopolymers. 50 (1): 13–22. doi: 10.1002/(sici)1097-0282(199907)50:1<13::aid-bip2>3.0.co;2-k. ISSN 0006-3525.
- ^ Sagermann, Martin; Mårtensson, Lars-Göran; Baase, Walter A.; Matthews, Brian W. (2002-03-01). "A test of proposed rules for helix capping: Implications for protein design". Protein Science. 11 (3): 516–521. doi: 10.1110/ps.39802. ISSN 1469-896X. PMC 2373482. PMID 11847274.
- ^ a b Leader, DP; Milner-White, EJ (2011). "The structure of the ends of alpha-helices in globular proteins". Proteins. 79 (3): 1010–1019. doi: 10.1002/prot.22942. PMID 21287629.
- ^ Milner-White, E. James; Nissink, J. Willem M.; Allen, Frank H.; Duddy, William J. (2004-11-01). "Recurring main-chain anion-binding motifs in short polypeptides: nests". Acta Crystallographica Section D. 60 (11): 1935–1942. doi: 10.1107/S0907444904021390. ISSN 0907-4449. PMID 15502299.
- ^ Datta, S; Uma, MV (1999). "Stereochemistry of Schellman motifs in peptides. Crystal structure of a hexapeptide with a C-terminal 6->1 hydrogen bond" (PDF). Biopolymers. 50 (1): 13–22. doi: 10.1002/(sici)1097-0282(199907)50:1<13::aid-bip2>3.0.co;2-k.
- ^ Watson, James D; Milner-White, E James (2002-01-11). "A novel main-chain anion-binding site in proteins: the nest. A particular combination of phi,psi values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions". Journal of Molecular Biology. 315 (2): 171–182. doi: 10.1006/jmbi.2001.5227. ISSN 0022-2836. PMID 11779237.
- ^ Leader, DP; Milner-White (2009). "Motivated Proteins: A web application for studying small three-dimensional protein motifs". BMC Bioinformatics. 10 (1): 60. doi: 10.1186/1471-2105-10-60. PMC 2651126. PMID 19210785.
- ^ Golovin, A; Henrick (2008). "MSDmotif: exploring protein sites and motifs". BMC Bioinformatics. 9 (1): 312. doi: 10.1186/1471-2105-9-312. PMC 2491636. PMID 18637174.
- ^ Newell, Nicholas E (2011-12-15). "Cascade detection for the extraction of localized sequence features; specificity results for HIV-1 protease and structure-function results for the Schellman loop". Bioinformatics. 27 (24): 3415–3422. doi: 10.1093/bioinformatics/btr594. ISSN 1367-4811. PMID 22039211.
- ^ Afzal, AM; Al-Shubailly, F (2014). "Bridging of anions by hydrogen bonds in nest motifs and its significance for Schellman loops and other larger motifs within proteins". Proteins. 82 (11): 3023–3031. doi: 10.1002/prot.24663. PMID 25132631.
External links

Schellman loops (also called Schellman motifs or paperclips) [1] [2] [3] [4] [5] [6] [7] [8] [9] [10] are commonly occurring structural features of proteins and polypeptides. [11] Each has six amino acid residues (labelled residues i to i+5) with two specific inter-mainchain hydrogen bonds (as in lower figure, i) and a characteristic main chain dihedral angle conformation. The CO group of residue i is hydrogen-bonded to the NH of residue i+5 (colored orange in upper figure), and the CO group of residue i+1 is hydrogen-bonded to the NH of residue i+4 ( beta turn, colored purple). Residues i+1, i+2, and i+3 have negative φ (phi) angle values and the phi value of residue i+4 is positive. Schellman loops incorporate a three amino acid residue RL nest (protein structural motif), [12] [13] in which three mainchain NH groups (from Schellman loop residues i+3 to i+5) form a concavity for hydrogen bonding to carbonyl oxygens. About 2.5% of amino acids in proteins belong to Schellman loops. Two websites are available for examining small motifs in proteins, Motivated Proteins: [1]; [14] or PDBeMotif: [2]. [15]
The majority of Schellman loops (82%) occur at the C-terminus of an alpha-helix such that residues i, i+1, i+2 and i+3 are part of the helix. Over a quarter of helices (28%) have a C-terminal Schellman loop. [10]
Occasional Schellman loops occur with seven instead of six residues. In these, the CO group of residue i is hydrogen-bonded to the NH of residue i+6, and the CO group of residue i+1 is hydrogen-bonded to the NH of residue i+5. Rare “left-handed” six-residue Schellman loops occur; these have the same hydrogen bonds, but residues i+1, i+2, and i+3 have positive φ values while the φ value of residue i+4 is negative; the nest is of the LR, rather than the RL, kind.
Amino acid propensities for the residues of the common type of Schellman loop have been described. [16] Residue i+4 is the one most-highly conserved; it has positive φ values; 70% of amino acids are glycine and none are proline.

Consideration of the hydrogen bonding in the nests of Schellman loops bound to mainchain oxygens reveals two main types of arrangement: 1,3-bridged or not. In one (lower figure, ii) the first and third nest NH groups are bridged by an oxygen atom. In the other (lower figure, iv) the first NH group is hydrogen bonded to the CO group of an amino acid four residues behind in the sequence, and none of the nest NH groups are bridged. [17] It seems that Schellman loops are less homogeneous than might have been expected.
The original Schellman criteria [1] result in the inclusion of features not now regarded as Schellman loops. A newer set of criteria is given in the first paragraph.
References
- ^ a b Schellman, C (1980). Protein Folding. Amsterdam: Elsevier. pp. 53–61.
- ^ Milner-White, E J (1988-02-05). "Recurring loop motif in proteins that occurs in right-handed and left-handed forms. Its relationship with alpha-helices and beta-bulge loops". Journal of Molecular Biology. 199 (3): 503–511. doi: 10.1016/0022-2836(88)90621-3. ISSN 0022-2836. PMID 3351939.
- ^ Aurora, R; Srinivasan, R; Rose, G D (1994). "Rules for alpha-helix termination by glycine" (PDF). Science. 264 (5162): 1126–1130. doi: 10.1126/science.8178170. ISSN 0036-8075. PMID 8178170.
- ^ Viguera, Ana Rosa; Serrano, Luis (1995-08-04). "Experimental Analysis of the Schellman Motif". Journal of Molecular Biology. 251 (1): 150–160. doi: 10.1006/jmbi.1995.0422. ISSN 0022-2836. PMID 7643384.
- ^ Aurora, R; Rose, G D (January 1998). "Helix capping". Protein Science. 7 (1): 21–38. doi: 10.1002/pro.5560070103. ISSN 0961-8368. PMC 2143812. PMID 9514257.
- ^ Kallenbach, Neville R.; Gong, Youxiang (January 1999). "C-Terminal capping motifs in model helical peptides". Bioorganic & Medicinal Chemistry. 7 (1): 143–151. doi: 10.1016/S0968-0896(98)00231-4. ISSN 0968-0896. PMID 10199664.
- ^ Sukumar, Muppalla; Gierasch, Lila M (August 1997). "Local interactions in a Schellman motif dictate interhelical arrangement in a protein fragment". Folding & Design. 2 (4): 211–222. doi: 10.1016/S1359-0278(97)00030-8. ISSN 1359-0278. PMID 9269562.
- ^ Datta, Saumen; Uma, Manjappara V.; Shamala, N.; Balaram, P. (1999). "Stereochemistry of Schellman motifs in peptides: crystal structure of a hexapeptide with a C-terminus 6→1 hydrogen bond" (PDF). Biopolymers. 50 (1): 13–22. doi: 10.1002/(sici)1097-0282(199907)50:1<13::aid-bip2>3.0.co;2-k. ISSN 0006-3525.
- ^ Sagermann, Martin; Mårtensson, Lars-Göran; Baase, Walter A.; Matthews, Brian W. (2002-03-01). "A test of proposed rules for helix capping: Implications for protein design". Protein Science. 11 (3): 516–521. doi: 10.1110/ps.39802. ISSN 1469-896X. PMC 2373482. PMID 11847274.
- ^ a b Leader, DP; Milner-White, EJ (2011). "The structure of the ends of alpha-helices in globular proteins". Proteins. 79 (3): 1010–1019. doi: 10.1002/prot.22942. PMID 21287629.
- ^ Milner-White, E. James; Nissink, J. Willem M.; Allen, Frank H.; Duddy, William J. (2004-11-01). "Recurring main-chain anion-binding motifs in short polypeptides: nests". Acta Crystallographica Section D. 60 (11): 1935–1942. doi: 10.1107/S0907444904021390. ISSN 0907-4449. PMID 15502299.
- ^ Datta, S; Uma, MV (1999). "Stereochemistry of Schellman motifs in peptides. Crystal structure of a hexapeptide with a C-terminal 6->1 hydrogen bond" (PDF). Biopolymers. 50 (1): 13–22. doi: 10.1002/(sici)1097-0282(199907)50:1<13::aid-bip2>3.0.co;2-k.
- ^ Watson, James D; Milner-White, E James (2002-01-11). "A novel main-chain anion-binding site in proteins: the nest. A particular combination of phi,psi values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions". Journal of Molecular Biology. 315 (2): 171–182. doi: 10.1006/jmbi.2001.5227. ISSN 0022-2836. PMID 11779237.
- ^ Leader, DP; Milner-White (2009). "Motivated Proteins: A web application for studying small three-dimensional protein motifs". BMC Bioinformatics. 10 (1): 60. doi: 10.1186/1471-2105-10-60. PMC 2651126. PMID 19210785.
- ^ Golovin, A; Henrick (2008). "MSDmotif: exploring protein sites and motifs". BMC Bioinformatics. 9 (1): 312. doi: 10.1186/1471-2105-9-312. PMC 2491636. PMID 18637174.
- ^ Newell, Nicholas E (2011-12-15). "Cascade detection for the extraction of localized sequence features; specificity results for HIV-1 protease and structure-function results for the Schellman loop". Bioinformatics. 27 (24): 3415–3422. doi: 10.1093/bioinformatics/btr594. ISSN 1367-4811. PMID 22039211.
- ^ Afzal, AM; Al-Shubailly, F (2014). "Bridging of anions by hydrogen bonds in nest motifs and its significance for Schellman loops and other larger motifs within proteins". Proteins. 82 (11): 3023–3031. doi: 10.1002/prot.24663. PMID 25132631.