| Developer(s) | Bioinformatics Solutions Inc |
|---|---|
| Stable release | PEAKS 8.5
/ October 12, 2017 |
| Operating system | Windows |
| Type | Mass Spec Protein Identification, De Novo Sequencing, Database Search & Quantification |
| License | Proprietary commercial software |
| Website | http://www.bioinfor.com/ |
PEAKS is a proteomics software program for tandem mass spectrometry designed for peptide sequencing, protein identification and quantification.
PEAKS is commonly used for peptide identification (Protein ID) through de novo peptide sequencing assisted search engine database searching. [1] PEAKS has also integrated PTM and mutation characterization through automatic peptide sequence tag based searching (SPIDER) [2] and PTM Identification. [3]
PEAKS provides a complete sequence for each peptide, confidence scores on individual amino acid assignments, simple reporting for high-throughput analysis, amongst other information.
The software has the ability to compare results of multiple search engines. PEAKS inChorus will cross check test results automatically with other protein ID search engines, like Sequest, OMSSA, X!Tandem and Mascot. This approach guards against false positive peptide assignments.
PEAKS Q is an add-on tool for protein quantification, supporting label ( ICAT, iTRAQ, SILAC, TMT, 018, etc.) and label free techniques.
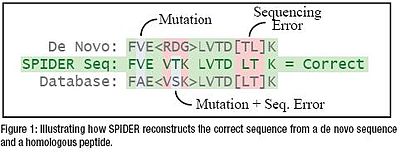
SPIDER is a sequence tag based search tool within PEAKS, which deals with the possible overlaps between the de novo sequencing errors and the homology mutations. It reconstructs the real peptide sequence by combining both the de novo sequence tag and the homolog, automatically and efficiently. [2]
A collection of algorithms used within the PEAKS software have been adapted and configured into a specialized project, PEAKS AB, which has proven to be the first method for automatic monoclonal antibody sequencing. [4]
- ^ Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.; Ma, B. (2011). "PEAKS DB: De Novo Sequencing Assisted Database Search for Sensitive and Accurate Peptide Identification". Molecular & Cellular Proteomics. 11 (4): M111.010587. doi: 10.1074/mcp.M111.010587. PMC 3322562. PMID 22186715.
- ^ a b Ma B, Johnson. De Novo sequencing and homology searching Molecular & Cellular Proteomics. 10.1074/mcp.O111.014902 (2011).
- ^ Han, X.; He, L.; Xin, L.; Shan, B.; Ma, B. (2011). "PEAKS PTM: Mass Spectrometry Based Identification of Peptides with Unspecified Modifications". Journal of Proteome Research. 10 (7): 2930–2936. doi: 10.1021/pr200153k. PMID 21609001.
- ^ Tran, Ngoc Hieu; Rahman, M. Ziaur; He, Lin; Xin, Lei; Shan, Baozhen; Li, Ming (26 August 2016). "Complete De Novo Assembly of Monoclonal Antibody Sequences". Scientific Reports. 6: 31730. Bibcode: 2016NatSR...631730T. doi: 10.1038/srep31730. ISSN 2045-2322. PMC 4999880. PMID 27562653.
| Developer(s) | Bioinformatics Solutions Inc |
|---|---|
| Stable release | PEAKS 8.5
/ October 12, 2017 |
| Operating system | Windows |
| Type | Mass Spec Protein Identification, De Novo Sequencing, Database Search & Quantification |
| License | Proprietary commercial software |
| Website | http://www.bioinfor.com/ |
PEAKS is a proteomics software program for tandem mass spectrometry designed for peptide sequencing, protein identification and quantification.
PEAKS is commonly used for peptide identification (Protein ID) through de novo peptide sequencing assisted search engine database searching. [1] PEAKS has also integrated PTM and mutation characterization through automatic peptide sequence tag based searching (SPIDER) [2] and PTM Identification. [3]
PEAKS provides a complete sequence for each peptide, confidence scores on individual amino acid assignments, simple reporting for high-throughput analysis, amongst other information.
The software has the ability to compare results of multiple search engines. PEAKS inChorus will cross check test results automatically with other protein ID search engines, like Sequest, OMSSA, X!Tandem and Mascot. This approach guards against false positive peptide assignments.
PEAKS Q is an add-on tool for protein quantification, supporting label ( ICAT, iTRAQ, SILAC, TMT, 018, etc.) and label free techniques.
SPIDER is a sequence tag based search tool within PEAKS, which deals with the possible overlaps between the de novo sequencing errors and the homology mutations. It reconstructs the real peptide sequence by combining both the de novo sequence tag and the homolog, automatically and efficiently. [2]

A collection of algorithms used within the PEAKS software have been adapted and configured into a specialized project, PEAKS AB, which has proven to be the first method for automatic monoclonal antibody sequencing. [4]
- ^ Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.; Ma, B. (2011). "PEAKS DB: De Novo Sequencing Assisted Database Search for Sensitive and Accurate Peptide Identification". Molecular & Cellular Proteomics. 11 (4): M111.010587. doi: 10.1074/mcp.M111.010587. PMC 3322562. PMID 22186715.
- ^ a b Ma B, Johnson. De Novo sequencing and homology searching Molecular & Cellular Proteomics. 10.1074/mcp.O111.014902 (2011).
- ^ Han, X.; He, L.; Xin, L.; Shan, B.; Ma, B. (2011). "PEAKS PTM: Mass Spectrometry Based Identification of Peptides with Unspecified Modifications". Journal of Proteome Research. 10 (7): 2930–2936. doi: 10.1021/pr200153k. PMID 21609001.
- ^ Tran, Ngoc Hieu; Rahman, M. Ziaur; He, Lin; Xin, Lei; Shan, Baozhen; Li, Ming (26 August 2016). "Complete De Novo Assembly of Monoclonal Antibody Sequences". Scientific Reports. 6: 31730. Bibcode: 2016NatSR...631730T. doi: 10.1038/srep31730. ISSN 2045-2322. PMC 4999880. PMID 27562653.